Abstract
Introduction
Acute myeloid leukemia (AML) is a highly heterogeneous disease with various genetic abnormalities. Cytogenetics, gene mutations, gene expression profiles, and microRNA expressions affect prognosis in AML patients. Some of these exams are cumbersome and not readily available. In contrast, routine complete blood counting and morphological classification is a simple and real time method, and may be used for prediction of response to treatment. Rapid peripheral blood (PB) blasts clearance after induction chemotherapy has been shown to predict prognosis in AML patients, but those studies recruited small cohorts or lacked molecular correlations. In this study, we aimed to not only investigate the prognostic significance of this method in a large cohort but also extend its implications to genetic and biological levels.
Method
A cohort of 333 de novo non-M3 AML patients who received standard chemotherapy at the National Taiwan University Hospital from 2003 to 2015 were included. The percentages of their PB blasts in the first week after initiation of induction chemotherapy were examined by morphological observation. Mutation analyses of 15 relevant molecular markers, including FLT3 (ITD and TKD), N-RAS, K-RAS, KIT, CEBPA, RUNX1, as well as NPM1, WT1, ASXL1, MLL/PTD, DNMT3A, IDH1, IDH2, and TET2, were performed in bone marrow mononuclear cells from 165 patients at diagnosis, who had available cryopreserved samples. We profiled whole-genome gene expression using Illumina HumanHT-12 v4 Expression BeadChip (Illumina, San Diego, CA) in 83 patients, who had enough mRNA for analyses. The difference in clinical features, cytogenetics, gene mutations, gene expression profiles, and treatment outcomes between patients who had PB blasts clearance within 7 days and those who failed were analyzed.
Results
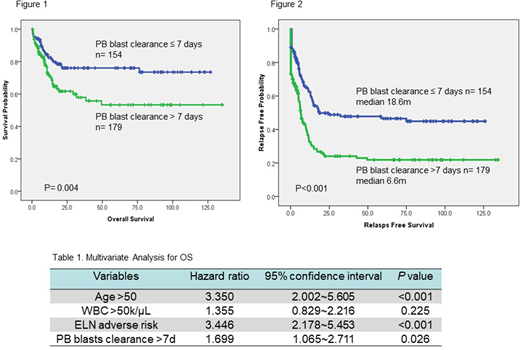
Among the 333 patients, 154 (46.2%) patients achieved clearance of PB blasts within 7 days of starting induction chemotherapy. There was no difference in gender, age, and hemograms at diagnosis between the two groups. The patients failed to clear PB blasts within one week had higher incidence of FAB M0 (5.6% vs. 0.6%, p=0.012), higher percentage of 2017 European LeukemiaNet (ELN) adverse-risk category (40.8% vs. 26.0%, p=0.004), and lower incidence of mutated NPM1 (10.6% vs. 22.5%, p=0.039), FLT3-ITD (11.8% vs. 26.6%, p=0.015), and biallelic mutated CEBPA (9.8% vs. 24.1%, p=0.015) than those who cleared the PB blasts in 7 days. Failure to clear PB blasts within 7 days was a significant unfavorable risk factor for overall survival (OS) [5-year survival rate 53.3% vs. 76.0%, p=0.004 (figure 1)], and relapse-free survival (RFS) [median RFS 6.6 months vs. 18.6 month, p<0.001 (figure 2)]. We then combined age, white blood cell count at diagnosis, ELN risk category, and clearance of PB blasts within 7 days as co-variables for a Cox regression proportional hazard model and found that failure to achieve clearance of PB blasts independently predicted shorter OS and RFS [HR=1.699, 95% CI 1.065-2.711, p=0.026 (table 1), and HR=1.896, 95% CI 1.398-2.573, p<0.001 respectively].
We profiled the global mRNA expression to explore the possible underlying molecular pathways which distinguish these two groups of patients. We identified 192 unique genes with differential expression (> 1.5 fold change, t-test p < 0.05). Gene-annotation enrichment analysis using DAVID 6.8 revealed that these differentially expressed genes were associated with abundant biological functions in a variety of categories. Among them were genes that were associated with cell proliferation (p = 9.68 × 10−4), and apoptosis (p = 3.08 × 10−3). Gene Set Enrichment Analysis (GSEA) revealed significant enrichment in "genes up-regulated in hematopoietic stem cells compared to hematopoietic progenitor cells" (empirical p<0.001) and "predominant long-term growth and self-renewal phenotype" (empirical p<0.001) in the patients with failure to clear PB blasts within 7 days.
Conclusion
This study demonstrated that rapid clearance of PB blasts is an independent favorable prognostic factor irrespective of age, WBC and 2017 ELN risk classification, and would be a simple and real time tool to predict the patients' prognosis. This parameter separates the patients into groups with distinctive clinical and biological features. Further prospective and larger cohorts are needed to confirm our observations.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal